Confocal laser scanning microscopy (CLSM or LSCM) is a technique for obtaining high-resolution optical images. The key feature of confocal microscopy is its ability to produce in-focus images of thick specimens, a process known as optical sectioning. Images are acquired point-by-point and reconstructed with a computer, allowing three-dimensional reconstructions of topologically-complex objects.

DESCRIPTION
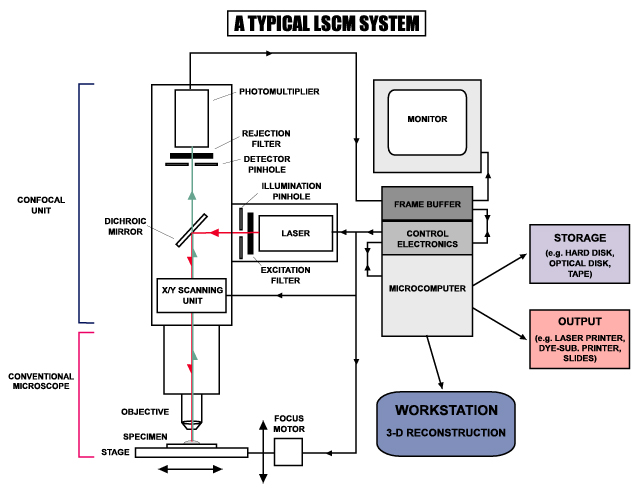
In a confocal laser scanning microscope, a laser beam passes through a light source aperture and then is focused by an objective lens into a small (ideally diffraction limited) focal volume within a fluorescent specimen. A mixture of emitted fluorescent light as well as reflected laser light from the illuminated spot is then recollected by the objective lens. A beam splitter separates the light mixture by allowing only the laser light to pass through and reflecting the fluorescent light into the detection apparatus. After passing a pinhole, the fluorescent light is detected by a photodetection device (a photomultiplier tube (PMT) or avalanche photodiode), transforming the light signal into an electrical one that is recorded by a computer.
The detector aperture obstructs the light that is not coming from the focal point, as shown by the dotted gray line in the image. The out-of-focus light is suppressed: most of their returning light is blocked by the pinhole, resulting in sharper images than those from conventional fluorescence microscopy techniques, and permits one to obtain images of various z axis planes (also known as z stacks) of the sample.
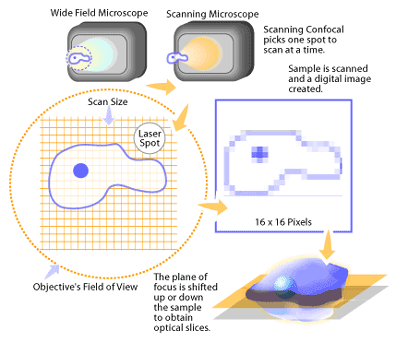
The detected light originating from an illuminated volume element within the specimen represents one pixel in the resulting image. As the laser scans over the plane of interest, a whole image is obtained pixel-by-pixel and line-by-line, whereas the brightness of a resulting image pixel corresponds to the relative intensity of detected fluorescent light. The beam is scanned across the sample in the horizontal plane by using one or more (servo controlled) oscillating mirrors. This scanning method usually has a low reaction latency and the scan speed can be varied. Slower scans provide a better signal-to-noise ratio, resulting in better contrast and higher resolution. Information can be collected from different focal planes by raising or lowering the microscope stage. The computer can generate a three-dimensional picture of a specimen by assembling a stack of these two-dimensional images from successive focal planes.

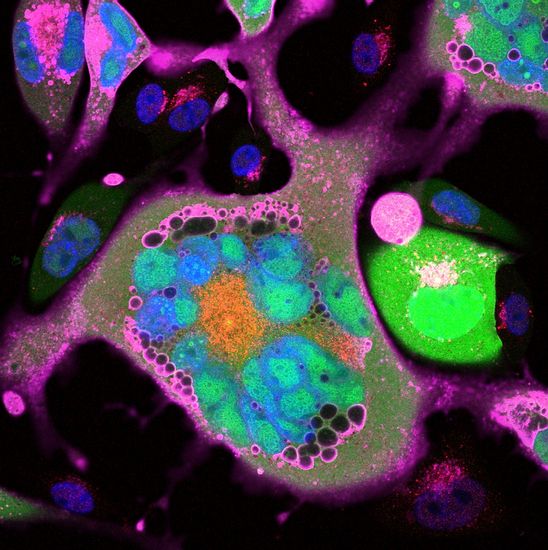
CONFOCAL IMAGE OF GFP FUSION PROTEIN

Confocal microscopy also provides a substantial improvement in lateral resolution and the capacity for direct, noninvasive, serial optical sectioning of intact, thick, living specimens with a minimum of sample preparation. Because CLSM depends on fluorescence, a sample usually needs to be treated with fluorescent dyes to make objects visible. However, the actual dye concentration can be low to minimize the disturbance of biological systems: some instruments can track single fluorescent molecules. Also, transgenic techniques can create organisms that produce their own fluorescent chimeric molecules (such as a fusion of GFP, green fluorescent protein with the protein of interest).
RESOLUTION OF IMAGES
CLSM is a scanning imaging technique in which the resolution obtained is best explained by comparing it with another scanning technique like that of the scanning electron microscope (SEM). Do not confuse CLSM with phonograph-like imaging—AFM or STM, for example, where the image is obtained by scanning with an atomic tip over a conducting surface.
In CLSM a fluorescent specimen is illuminated by a point laser source, and each volume element is associated with a discrete fluorescence intensity. Here, the size of the scanning volume is determined by the spot size (close to diffraction limit) of the optical system because the image of the scanning laser is not an infinitely small point but a three-dimensional diffraction pattern. The size of this diffraction pattern and the focal volume it defines is controlled by the numerical aperture of the system's objective lens and the wavelength of the laser used. This can be seen as the classical resolution limit of conventional optical microscopes using wide-field illumination. However, with confocal microscopy it is even possible to overcome this resolution limit of wide-field illuminating techniques because only light generated in a small volume element is detected at a given time. Here the effective volume of light generation is usually smaller than the volume of illumination; that is, the diffraction pattern of detectable light creation is sharper and smaller than the diffraction pattern of illumination. The resolution limit in confocal microscopy depends not only on the probability of illumination but also on the probability of creating enough detectable photons, so that the actual addressable volume being associated with a generated light intensity is smaller than the illuminated volume. Depending on the fluorescence properties of the used dyes, there is a more or less subtle improvement in lateral resolution compared to conventional microscopes. However, with light creation processes with much lower probabilities of occurrence such as second harmonic generation (SHG), the volume of addressing is reduced to a small region of highest laser illumination intensity, substantially improving lateral resolution. Unfortunately, the probability decrease in creation of detectable photons negatively affects the signal-to-noise ratio. One can compensate for this effect by using more sensitive photodetectors or by increasing the intensity of the illuminating laser point source. Increasing the intensity of illumination later risks excessive bleaching or other damage to the specimen of interest, especially for experiments in which comparison of fluorescence brightness is required.
BIOMEDICAL APPLICATIONS
1.)CLSM is widely-used in numerous biological science disciplines, from cell biology and genetics to microbiology and developmental biology.

(Organization of actin arrays in mitosis)
2.)Clinically, CLSM is used in the evaluation of various eye diseases, and is particularly useful for imaging, qualitative analysis, and quantification of endothelial cells of the cornea. It is used for localizing and identifying the presence of filamentary fungal elements in the corneal stroma in cases of keratomycosis, enabling rapid diagnosis and thereby early institution of definitive therapy.
3.)Research into CLSM techniques for endoscopic procedures is also showing promise.
4.)CLSM is also used as the data retrieval mechanism in some 3D optical data storage systems and
5.)It has helped determine the age of the Magdalen papyrus.